-
Notifications
You must be signed in to change notification settings - Fork 152
Fix issue 255 #273
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Fix issue 255 #273
Conversation
I was mistaken about this, sorry, correcting approach. |
|
@quinn-dougherty That looks perfect. Just fix the typo -> "I know the date of the first [...]" (remove 'number of') |
src/penn_chime/models.py
Outdated
| if p.n_days_since_first_hospitalized is not None: | ||
| # do not shorten this to `if foo:` form. if you do and n_days_since_first_hospitalized is 0, | ||
| # program will crash. | ||
| p.doubling_time = self.argmin_dt(np.linspace(1, 15, 29), p) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is a really horrifying thought, but is there a principled reason the doubling time can never be less than 1 day? Super-spreader events would seem to do exactly that.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
that's a hardcoded constant from Corey's notebook that we can factor into the settings.cfg file if we wish. since we're pushing to prod weekly, we only hope that things don't get that drastic too quickly ;)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
More broadly, if the parameter sweep from argmin_dt returns a value at either extreme of its range, it means either
- The best fit value is approximately the given limit
- The fit failed because the true result is outside the range
We need to not let the latter case go through without warning or examination. It may just as well be an indicator of mis-keyed inputs, as of the math honestly saying anything about the circumstances.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
There's a physical limit on doubling time down around 0.1, since that implies 10 daily doublings, or about 1000x, which exhausts itself pretty damn quickly. I think there's some threshold higher than that where we should warn the user that the computed value is implausibly low
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
i think you're right. i'll add in some sort of warning system.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I believe that a different merge allowed doubling time to be a float/fractional.
|
A UI note for as this progresses: The number of days for which the user asks for a projection needs to be in addition to days in the back-cast - i.e. the admissions and census charts should run from |
|
@cjbayesian This is not parity with prod. The codebase considers the entire how should we proceed? |
|
Yep, @PhilMiller is right that I had fixed the susceptible calculation in the notebook per issue #211 . |
#276 merged |
|
@PhilMiller and @quinn-dougherty : I have a coherent way to handle either case of user input (doubling_time or n_days_since_first_hospitalized) which addresses also #187 . I just pushed an update to my notebook showing how. It's basically the same fitting procedure, just holding |
|
Per Phil's comments, someone remind us to have a talk about what this |
pushing some commits without the 187 piggyback shortly |
sorry, this is gonna have to be another PR. racing to get the basics of 255 into Prod monday. |
|
TODO:
|
src/penn_chime/models.py
Outdated
| recovered, | ||
| beta, | ||
| gamma, | ||
| p.n_days |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Shouldn't this be p.n_days + p.n_days_since_first_hospitalization?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
all i'll say is that wouldn't be parity with the notebook. @cjbayesian
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
OK, so we'll fix this later then. We really want the raw_df to hold all of the data instead of having to bodge n_days_since... into every function.
I'll get a copy of the notebook and test for equivalent output later.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
However, this input can no longer be described as the number of days to project as stated in the ui. It is projecting less than n_days, it is projecting from the first infection. The ui / users here will be confusing / confused.
|
hi -- are we ok if i piggyback updating |
… into my_more_issue_255
Put social distancing effect back in. Starts at current day.
These are irrelevent to the calculations and only cause user confusion.
Remove confirmed cases & detection prob
Add time as a parameter

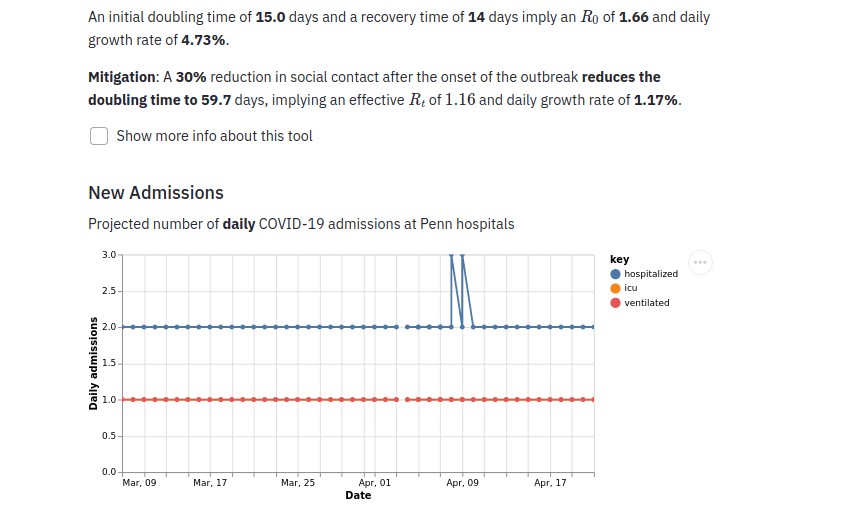
Closes #255
notebook for reference
found a clean way to do it in streamlit's UI that doesn't build up ugly technical debt.
TODO: need to rearrange the order in which computations happen in order to get it to actually work. in the notebook, inferred doubling time is a function of assumed doubling time, so i just need to make that work with autopopulating the form with a default (and not write myself into literal circles)